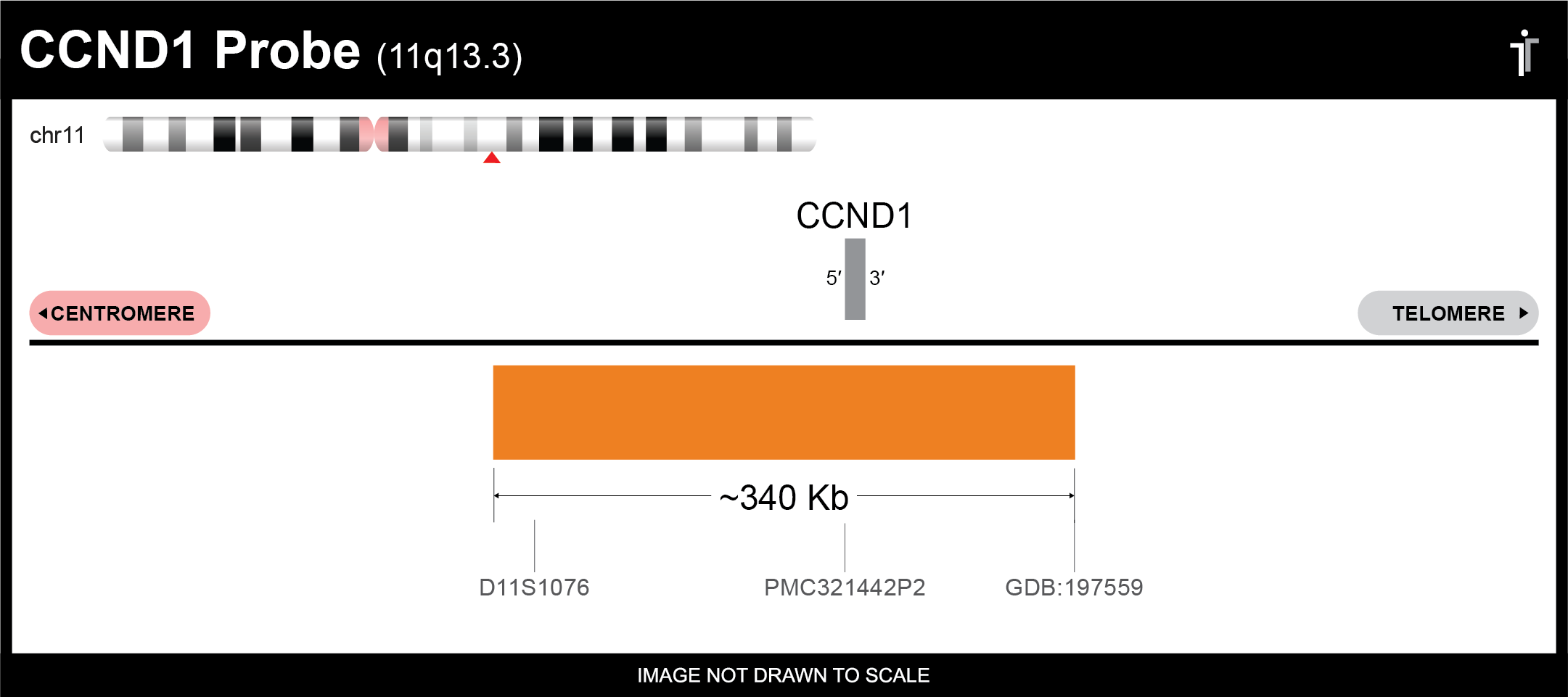
CCND1 FISH Probe
Our CCND1 probe is designed to detect CCND1 amplifications and deletions. The probe comes labeled in orange, but can be customized to meet your needs.
Gene Background: Cyclin D1 (CCND1) regulates G1 to S-phase cell cycle transition alongside its binding partners, CDK4 and CDK6, via phosphorylation and inactivation of the retinoblastoma protein (pRb).1 First identified in 1991, CCND1 has been found amplified in about 9-30% of breast cancer cases.1 It’s also overexpressed in melanoma, mantle cell lymphoma, glioma, and endometrial, bladder, brain, esophageal, and colorectal cancer.2
** This product is for in vitro and research use only. This product is not intended for diagnostic use.

| SKU | Test Kits | Buffer | Dye Color | Order Now |
|---|---|---|---|---|
| CCND1-20-OR (Standard Design) | 20 (40 μL) | 200 μL |

|
|
| CCND1-20-BIO | 20 (40 μL) | 200 μL |

|
|
| CCND1-20-RE | 20 (40 μL) | 200 μL |

|
|
| CCND1-20-DIG | 20 (40 μL) | 200 μL |

|
|
| CCND1-20-AQ | 20 (40 μL) | 200 μL |

|
|
| CCND1-20-GR | 20 (40 μL) | 200 μL |

|
|
| CCND1-20-GO | 20 (40 μL) | 200 μL |

|
Gene Summary
The protein encoded by this gene belongs to the highly conserved cyclin family, whose members are characterized by a dramatic periodicity in protein abundance throughout the cell cycle. Cyclins function as regulators of CDK kinases. Different cyclins exhibit distinct expression and degradation patterns which contribute to the temporal coordination of each mitotic event. This cyclin forms a complex with and functions as a regulatory subunit of CDK4 or CDK6, whose activity is required for cell cycle G1/S transition. This protein has been shown to interact with tumor suppressor protein Rb and the expression of this gene is regulated positively by Rb. Mutations, amplification and overexpression of this gene, which alters cell cycle progression, are observed frequently in a variety of tumors and may contribute to tumorigenesis. [provided by RefSeq, Jul 2008]
Gene Details
Gene Symbol: CCND1
Gene Name: Cyclin D1
Chromosome: CHR11: 69455872-69469242
Locus: 11q13.3
FISH Probe Protocols
| Protocol, Procedure, or Form Name | Last Modified | Download |
|---|